Physicists describe genome ejection by bacteriophage viruses
15 March 2013
Bacteriophages, or phages in short, are viruses of bacteria. Sixty years ago the first experiments showed that nucleic acid is the major component of phage particles that is ejected into bacterial cells during infection. In light of the recent advances in electron microscopy, the structure, packaging and energetics of the DNA confined in a phage capsid are beginning to be better understood, but the ejection process still remains shrouded in mystery. Debabrata Panja (UU), who is also a guest at ITFA, has been active in this field for some years. Together with Ian J. Molineux, a phage virologist at the University of Texas at Austin, he has formulated a (physical) hydrodynamic model for the mechanism of phage genome ejection into bacterial cells.
Textbook knowledge to the test
It seems to be textbook knowledge: the phage binds to the specific receptors on the bacterial cell surface, drills a hole, and squirts its genome into the cell. The genome hijacks the cell’s machinery, and the rest is history. But it does not tell us where the energy to squirt the genome into the cell actually comes from.
A popular explanation is that the genome is highly condensed in the phage capsid. A typical phage capsid is about 60 nm in diameter, in which, for double-stranded DNA phages, typically 15 micron long DNA is packaged. The DNA remains coiled within the capsid “against its will”, pushing against the capsid resisting electrical repulsion forces among the negatively charged phosphates on its backbone, as well as resisting bending in tight circles. When the specific receptor on the bacterial cell surface opens the gate of the capsid, the genome flies out. At least, that is the prediction of the “continuum mechanics model”.
But there is a problem with this explanation. Calculation shows that the osmotic pressure within bacterial cells, commonly known as “turgor” – which bacteria must maintain at a value higher than that of the culture medium in order to grow – resists the coiled DNA’s will to fly out to the point that by itself only about 20-50% of the genome will enter the cell, and not more. Of course, one can think of “active” mechanisms to complete the rest of the genome entry into the cell, but such generic mechanisms are experimentally unsupported.
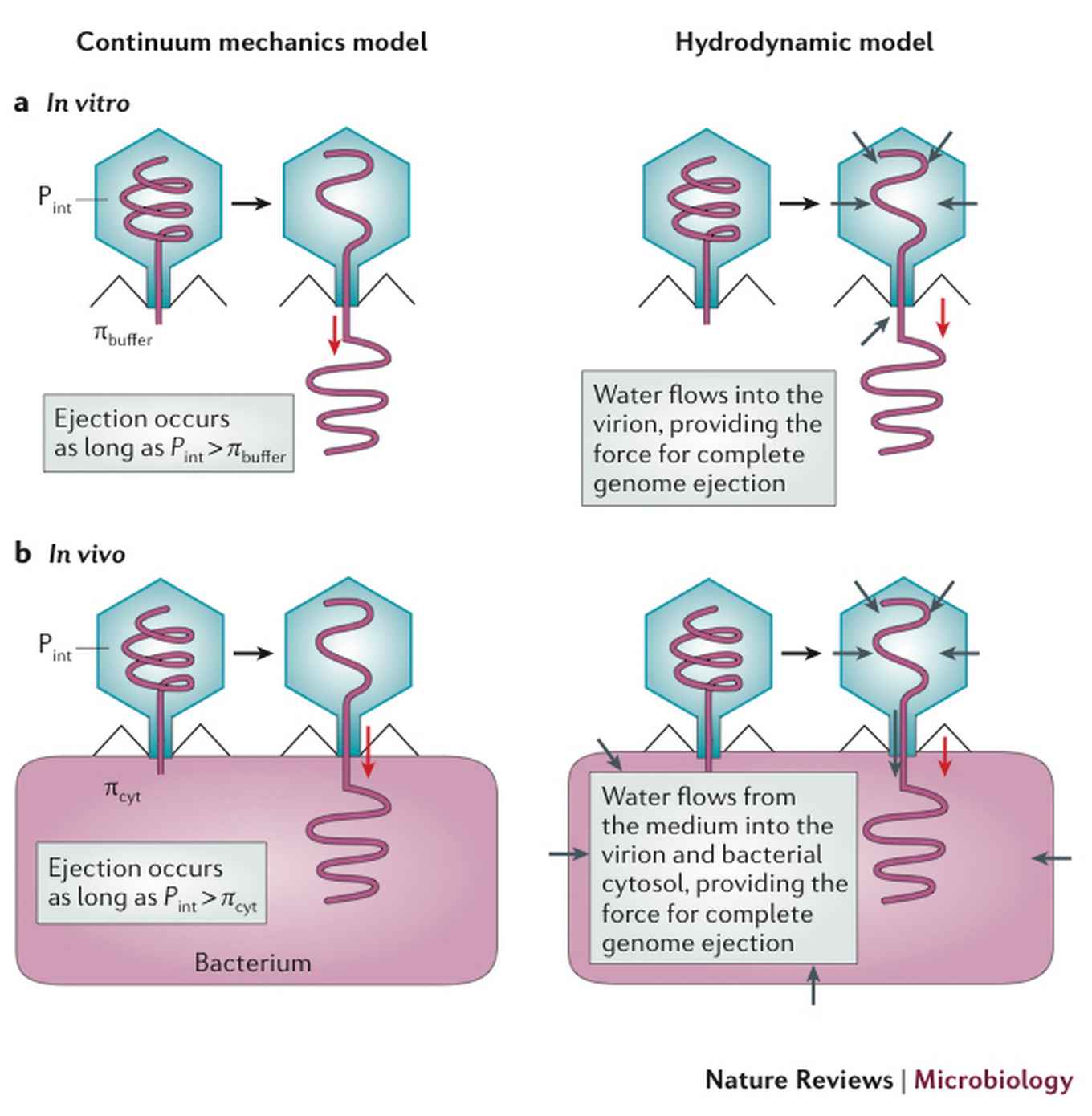
Hydrodynamic model: go with the flow
The authors proposed a hydrodynamic model that uses the higher value of the cells’ turgor in comparison to the culture medium. When the capsid gate is opened and simultaneously a hole is drilled on the bacterial cell surface, water will flow from the culture medium through the phage capsid and drain into the cell up the osmotic gradient. The contents of the capsid will be flushed by this water flow into the cell, much like logs floating down the river stream, and the compaction of the genome within the capsid is largely irrelevant in this process. Unlike the continuum mechanics model, the hydrodynamic model respects phage phenomenology, such as leakage of K+ ions from the bacterial cell into the culture medium during infection, and immunity of bacterial cells to phage infections in the so-called “stationary phase”.
Publication detail
Ian J. Molineux and Debabrata Panja:Popping the cork: mechanisms of phage genome ejection, Nature Reviews Microbiology 11, 194-204 (March 2013).